Fighting Fusarium Through Molecular Genetics
In Madison, Wisconsin, as part of the U.S. Wheat and Barley Scab Initiative (USWBSI), ARS plant molecular biologist Ron Skadsen is working to understand and improve barley’s resistance to the fungal pathogen Fusarium graminearum, which causes the devastating disease commonly known as “scab.” Scab reduces yield by causing sterility and shrunken kernels and contaminating the grain with mycotoxins.
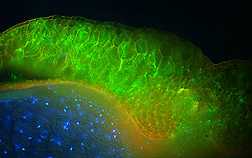
Skadsen, who is in the Cereal Crops Research Unit, first sought to identify the barley tissues that Fusarium most readily attacks. He infected barley seed spikes with Fusarium transformed to contain a green fluorescent protein that makes the fungus glow neon green when examined under a fluorescence microscope. Skadsen found that Fusarium attacks the protruding seed tip of the developing seed, the soft tissue connected with it (just under the hull), and, to a lesser extent, the seed’s outer hull.
Based on his findings, Skadsen’s team developed gene promoters that can be used to “turn on” genes that defend against Fusarium in these susceptible tissues. The promoters—from barley genes Lem1, Lem2, and Ltp6—are attached to barley antifungal genes to turn them on at specific locations.
“Knowing which parts of the barley plant Fusarium attacks gives us insight into how the infection process works,” says Skadsen. “We now know where to knock out Fusarium in the early stages of infection, which will aid targeted breeding and biotechnology strategies for making barley resistant.”
|
|
Previous studies examining Fusarium infection found that the fungus will liquefy the starchy part of the seed within 5 days after infection. But these studies used detached seeds. Skadsen found that, even 16 days after infection, Fusarium does not penetrate the starchy endosperm when the seed remains attached to the spike. This means breeders can focus on looking for traits that will prevent or head off the fungus’s early penetration.
Skadsen and research leader Cynthia Henson are also looking to understand the biochemistry of susceptible barley tissues through metabolic profiling during the first 3 days of infection. They found that there is a shift in sugar levels, especially the appearance of and rapid increase in the sugar alcohol ribitol, a metabolite that has not been extensively studied. The scientists are next looking to see whether the shifts in various sugar levels are caused by the plant’s mobilizing sugar away from the infection point or by the fungus’s taking specific sugars and metabolites away from the plant through feeding. They hope these results will inform researchers how Fusarium alters the metabolism of barley tissues to optimize its own nutrition.
|
|
Marker-Assisted Breeding of Scab-Resistant Barleys
Shiaoman Chao and her team at an ARS genotyping laboratory in Fargo, North Dakota, work closely with small-grains breeders in the Northern Plains region to use marker technologies to improve crops. Chao, a molecular geneticist, uses current genomics information to develop DNA markers tagged to important agronomic traits. Once appropriate markers are identified, they can be used in breeding populations to increase the efficiency of selection.
“At Fargo, we provide genotyping service to all small-grains breeders located in the region,” says Chao, who is in the ARS Cereal Crops Research Unit.
Rich Horsley at North Dakota State University (NDSU) and Kevin Smith at the University of Minnesota (UMN) are key collaborators in Chao’s work, providing breeding lines for the ARS Fargo team to do the genotyping.
To accelerate the rate of deployment of genes for resistance to scab using marker technologies, the Fargo lab has developed a sample preparation protocol and high-throughput genotyping procedures that are both efficient and cost effective for carrying out marker-assisted breeding.
|
|
Breeders send samples generated in their breeding programs by inserting leaf clippings into 96-well plates provided by Fargo lab. Much of the genotyping process has been semi-automated through use of robotic instruments.
High-throughput DNA extraction and marker genotyping protocols can help scientists conduct genetic mapping studies with large populations. This has been important in mapping scab resistance in barley because three of the most important resistance genes in barley are located very close to undesirable genes, which creates a substantial problem for breeders. But it can be overcome by screening very large populations with markers and identifying progeny in which the undesirable traits are unlinked from the resistance trait.
Chao screens breeding lines with resistance to scab using DNA markers previously identified as linked to the resistance genes in both wheat and barley. She also works on DNA markers for other traits, such as protein quality and resistance to leaf rust and tan spot in wheat and resistance to net blotch and Septoria speckled leaf blotch in barley.
“Creation of the ARS genotyping centers has dramatically changed the way small-grains breeders think about using DNA markers in breeding,” says Smith, who runs the barley breeding project at UMN. “There, the genotyping is done at a scale and speed that would not be possible if left to individual breeding programs.”
USWBSI Projects in North Dakota Take Shape
There are a total of 145 USWBSI projects in all research categories, according to the initiative’s manager, Sue Canty. Among USWBSI’s top objectives is the reduction of mycotoxins. This is important to the producers, processors, and consumers of wheat and barley.
Rich Horsley, a professor and barley breeder at NDSU, has been using his USWBSI grant for development of improved six-rowed and two-rowed germplasm. Horsley is also coordinating a Fusarium head blight (FHB) nursery at Zhejiang University in Hangzhou, China. Highlights include advancement of the six-rowed barley breeding line ND20448 into the final stages of plant-scale malting and brewing evaluation by the American Malting Barley Association, Inc. (AMBA).
Horsley has collaborated with Chao on mapping quantitative trait loci that confer resistance to scab and reduce accumulation of the mycotoxin known as “deoxynivalenol.” Identification of markers useful for screening across the multiple pedigrees and genetic backgrounds used by the NDSU barley breeding program has been unrealized until 2009. In the past, markers would work in some populations and not others. Using markers identified in research conducted by collaborators at UMN, Chao was able to genotype all six-rowed lines used as parents in crosses the past 4 years and all six-rowed lines grown in yield trials in the summer of 2009.
Data provided by Chao has allowed Horsley’s program to be more efficient in selecting lines as parents for this fall’s crossing block, planning which crosses to make, and determining which lines should be candidates for advancement to 2010’s yield trials. The project’s goal for 2010 is to submit to Chao’s laboratory leaf tissue from early-generation lines so the research team can determine which lines to advance for further testing in the breeding program.
Scab is a major factor in the decline of malting barley production to historic lows in the Dakotas and Minnesota, key states for raw material for the U.S. malting and brewing industry. “Research outputs from the USWBSI are expected to help address the production decline, reducing the need to source malting barley long distances,” says AMBA president Mike Davis.
CAP Program Key to Developing Elite Barley Germplasm
The focus of Coordinated Agricultural Project (CAP) research is to identify molecular markers that will speed up barley breeding efforts. The novelty of this approach is that mapping of important traits is done using contemporary breeding populations. The result is that breeding and mapping are done in parallel, accelerating translation of genetic information useful to plant breeding. Begun in 2005, CAP has already identified genetic markers associated with malting quality traits, winter hardiness, and resistance to scab. (For more information about ARS’s malting-quality work, see the story on page 7 of this issue.)
The goal of CAP is to detect single-nucleotide polymorphism (SNP) variations located at 3,072 different positions in the genomes of 3,840 barley breeding lines studied over a 4-year period. To detect these genetic variations, the Fargo lab is using a high-throughput genotyping system—a speedy and efficient way of analyzing SNPs—that’s capable of analyzing SNPs located at 1,536 positions of 96 individuals in a single reaction assay.
Chao is ARS’s lead scientist in CAP. Her team in Fargo carries out the high-throughput genotyping to generate 10 million data points. CAP participants mine this data and identify markers for barley improvement. The results have been published in scientific papers on evaluating the genetic diversity present among U.S. barley breeding programs. All findings in identifying SNP markers closely associated with various agronomic traits, such as scab resistance, are made publicly available to assist all barley breeders.—By Alfredo Flores and Stephanie Yao, Agricultural Research Service Information Staff.
This research is part of Plant Diseases (#303), Plant Biological and Molecular Processes (#302), and Plant Genetic Resources, Genomics, and Genetic Improvement (#301), three ARS national programs described at www.nps.ars.usda.gov.
To reach scientists mentioned in this article, contact Alfredo Flores, USDA-ARS Information Staff, 5601 Sunnyside Ave., Beltsville, MD 20705-5129; (301) 504-1627.
U.S. Wheat and Barley Scab Initiative and Barley Coordinated Agricultural Project
Protecting barley from disease and developing improved varieties involves coordinated USDA research supported by both intramural and competitive grants programs. As a prime example, ARS manages the U.S. Wheat and Barley Scab Initiative (USWBSI), targeted to combating the most devastating disease of barley and wheat in North America. ARS has joined with researchers, producers, millers, and processors to develop a pragmatic action plan to minimize the threat of Fusarium head blight (FHB), sometimes called “scab.”
A major goal of the USWBSI is to develop FHB-resistant barley varieties with good malting quality. ARS intramural researchers are partnering with more than 75 scientists receiving USWBSI competitive grant awards from 22 universities and international organizations in coordinated efforts to combat FHB and advance food safety and security. The action plan and more information on the USWBSI can be found at scabusa.org.
Barley breeding is being advanced by the Barley Coordinated Agricultural Project (CAP), supported by USDA’s Agriculture Research Funding Initiative, a collaborative effort of 30 scientists from 19 institutions, including ARS, that leverages genomics information and technology to advance barley breeding. Research in barley genetics/genomics, breeding, pathology, food science, and statistics makes up CAP. More information is available at barleycap.org.
"Fighting Fusarium Through Molecular Genetics" was published in the February 2010 issue of Agricultural Research magazine.