Campylobacter: Unmasking the
|
|
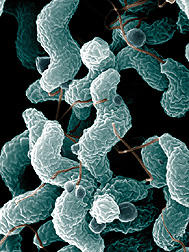 Campylobacter bacteria are the number-one cause of food-related gastrointestinal illness in the United States. To learn more about this pathogen, ARS scientists are sequencing multiple Campylobacter genomes. This scanning electron microscope image shows the characteristic spiral, or corkscrew, shape of C. jejuni cells and related structures. (K11505-1) |
The "juice" that always seems to leak out of those packages of fresh chicken you bring home from the supermarket can make a big mess on your kitchen counter. But more importantly, the juice can pose a hazard to your health. Nasty microbes called Campylobacter jejuni can live in that liquid and on the skin of fresh, uncooked poultry. Thoroughly cooking chicken—by grilling, frying, roasting, or baking—kills this food-poisoning microbe. But if you accidentally splash some of the raw juice on food that you'd planned to eat uncooked, such as leafy greens for a fresh salad, you'd be wise to throw them out. Here's why: If the microbe takes hold on those greens, as it is very adept at doing, you might be in for a case of campylobacteriosis food poisoning that you won't soon forget. Campylobacter is thought to be the leading cause of bacterial food poisoning in humans and is likely the perpetrator of more than 400 million cases of diarrhea every year. Though being careful when you handle raw poultry should help keep you safe, ARS researchers want to do more to zap this microbial menace before it reaches your home. |
 Microarrays, or gene chips, enable scientists to get a quick look at thousands of genes in a single experiment. Here, technician Sharon Horn monitors robotic equipment as it imprints Campylobacter microarrays on glass slides. (K11465-1) |
At Albany, California, scientists in the ARS Produce Safety and Microbiology Research Unit are making key advances in the international effort to clobber Campylobacter. The California team, based at the Western Regional Research Center, is focusing on Campylobacter's genes. Why the interest in the microbe's genetic makeup? Because investigating genes may lead to discovery of faster, more reliable ways to detect the microbe in samples from humans and other animals, food, and water. In addition, gene-based research opens the door to simpler, less-expensive tactics for distinguishing look-alike species and strains of Campylobacter and its close relatives, such as the Arcobacters. This will enable experts to quickly finger culprit microbes in food poisoning outbreaks. Finally, the studies may lead to innovative, environmentally friendly techniques to circumvent the genes that make C. jejuni strains so successful in causing human gastrointestinal upset and in some cases paralysis or even death. |
 Technician Guilin Wang sets the conditions for operation of an automated robotic system for purifying DNA. High-quality DNA is required for spotting onto glass slides for microarray experiments. (K11476-1) |
Working with the Institute for Genomic Research, Rockville, Maryland, the Albany scientists have decoded the makeup, or sequence, of all the genes and other genetic material in a specially selected strain of C. jejuni. This research represents the first time that a C. jejuni strain from a farm animal—in this case, a market chicken—has been sequenced. That's important, notes research leader Robert E. Mandrell, because chicken is the leading source of the bacterium in food. Earlier C. jejuni genome sequencing, performed elsewhere, was based on a specimen from a gastroenteritis patient and was lacking key features, such as the ability to colonize chickens, Mandrell says. The next step: Zero in on specific genes. "We're particularly interested in the genes that make Campylobacter so viable and virulent," says ARS molecular biologist William G. Miller. They're targeting, for instance, genes that carry the code for making oligosaccharides. These compounds likely enable the microbe to stick like glue to chicken skin in the poultry processing plant even though the birds are bathed and rinsed with chlorinated water. The oligosaccharides might be important in invading and colonizing the human body, as well, Miller notes. |
 Technician Sharon Horn and microbiologist William Miller prepare samples of Campylobacter for automated analysis of the structure, or sequence, of the DNA. The colored peaks on the computer screen show the sequence of a DNA sample from an earlier run. (K11472-1) |
With this genome sequence information in hand, the scientists are developing microarrays, or gene chips, that make possible a quick look at thousands of genes in a single experiment. For these analyses, robotic equipment precisely places pieces of the pathogen's DNA in an array of infinitesimally small droplets on glass microscope slides. "We build and use these microarrays to compare and contrast DNA of various Campylobacter samples," explains microbiologist Craig T. Parker. "We're also using microarrays to get a snapshot of genes in action so that we can see when genes are turned on or off." For example, Parker is pinpointing the genes that are active in helping Campylobacter overcome our bodies' protective actions. By tracking the action of the microbes' genes, Parker and co-investigators may be able to determine a way to derail them. |
 Research leader Robert Mandrell (left) and microbiologist Craig Parker, both of the Produce Safety and Microbiology Research Unit, examine an image of the results of a microarray experiment comparing over 1,700 genes of Campylobacter jejuni strains from farm animals and humans. (K11462-1) |
Though C. jejuni has grabbed center stage because of its known virulence, its relatives are also of interest. The Albany studies of C. coli, C. lari, and C. upsaliensis, for example, are attracting the attention of member nations in a three-continent collaboration called "Campycheck," formed to evaluate the importance of these lesser-known or newly emerging species. The Albany scientists and colleagues from the ARS Richard B. Russell Agricultural Research Center, Athens, Georgia, are advisors to Campycheck. In clinical laboratories, these less-studied pathogens may inadvertently be killed by the antibiotics used to identify the better-known ones. The likely result? An inaccurate picture of their prevalence and virulence. Campycheck may yield a detailed, accurate picture. The Campylobacter studies in the United States and abroad might never completely eliminate the need for careful handling of raw poultry in our homes or the kitchens of school cafeterias, fine restaurants, and other eateries. But the research can reduce our chances of ever encountering this unruly microbe.—By Marcia Wood, Agricultural Research Service Information Staff. This research is part of Food Safety, an ARS National Program (#108) described on the World Wide Web at www.nps.ars.usda.gov. To reach scientists mentioned in this story, contact Marcia Wood, USDA-ARS Information Staff, 5601 Sunnyside Ave., Beltsville, MD 20705-5129; phone (301) 504-1662, fax (301) 504-1641. What Makes a Campylobacter Strain Virulent? Here's the puzzle: You have two samples of what seem to be the food-poisoning microbe Campylobacter jejuni. A quick look at the specimens with a microarray assay (see main story) shows no immediately apparent differences in their genes. But when you expose piglets—animals susceptible to this microbe—to the bacteria, one strain makes the animals ill, while the other affects them only mildly. Why the difference? ARS food safety researchers Craig T. Parker, at Albany, California, and colleague Michael E. Konkel at Washington State University in Pullman, are designing a series of experiments that should enable them to find out. What's more, their work may help other scientists who are investigating the virulence of other major foodborne pathogens. Even though their preliminary microarray scan failed to reveal significant differences in the C. jejuni specimens' DNA, this technology offers another option—one that allows them to delve more deeply. Instead of beginning with the microbe's DNA, these followup assays begin with RNA—genetic material that's formed when the DNA, or genes, becomes active. In these tests, the scientists will place the two strains in petri dishes with colonies of a type of human intestinal cell. Called epithelial cells, they're the target of real-life Campylobacter attacks. The researchers will take samples of the two strains at successive intervals, looking for changes in RNA that occur over time. RNA extracted from the strains provides tell-tale evidence of genes that went into action. The work is much like that of police detectives who analyze evidence to reconstruct what really happened at a crime scene. The scientists use an enzyme called reverse transcriptase to match up the RNA to a version of the DNA from which it originated. Then, they use the microarray assay to discern the differences between that DNA and the microbe's DNA as it existed at the outset of the experiment. The comparison should reveal genes that were activated in the attack and genes that remained silent. In earlier work at Pullman, collaborator Konkel uncovered one such C. jejuni gene. Named ciaB, short for Campylobacter invasion antigen B, it cues the microbe to secrete a similarly named protein, CiaB, which apparently plays a crucial role in enabling the bacterium to penetrate epithelial cells. Though undoubtedly key to C. jejuni's invasions, it is unlikely to act alone. The West Coast scientists expect to uncover other genes that will lead them into the dark heart of Campylobacter's virulence.—By Marcia Wood, Agricultural Research Service Information Staff.
"Campylobacter: Unmasking the Secret Genes of a Food-Poisoning Culprit" was published in the October 2004 issue of Agricultural Research magazine.
|






