Detecting Deadly Colonies of E. coli
Shiga-toxin-producing Escherichia coli (STEC) serogroup O157:H7 has long been associated with severe forms of foodborne illnesses. But these bacterial bad guys have many cousins, and six other STEC groups have also been linked to serious illnesses and outbreaks of disease.
In 2011, the U.S. Department of Agriculture’s Food Safety and Inspection Service (FSIS) announced a regulation that, as of June 2012, certain beef products that test positive for any of these six groups, nicknamed the “Big Six,” would be banned from being sold for public consumption. As USDA’s regulatory arm, FSIS employs inspectors who collect meat samples on the front lines of meat-processing facilities. Those samples are then tested at FSIS laboratories for the presence of foodborne pathogens to help ensure that consumers’ food is safe before it is sold.
Identifying provenanalytical methods to detect other types of STEC besides O157:H7 in meat samples has always been a government priority. But in 2011, most of the existing detection methods for the Big Six were in the research stage and were not commercially available.
“Early laboratory methods to detect the Big Six were time-consuming and cost prohibitive,” says William Cray, chief of the microbiology branch of the USDA-FSIS Eastern Laboratory in Athens, Georgia. “Improved, validated methods were needed, and fast.”
FSIS scientists have long collaborated with researchers at the ARS Eastern Regional Research Center (ERRC) at Wyndmoor, Pennsylvania, so working together to produce a fast, accurate detection method for the Big Six was a natural progression.
ARS chemist Marjorie Medina and colleagues developed an analytical method that screened for all six non-O157 STEC groups. Medina—who is lead author of a published study about the method—recently retired from ERRC’s Residue Chemistry and Predictive Microbiology Unit. Coauthors include colleagues in ERRC’s Molecular Characterization and Foodborne Pathogens Unit; the ARS Biosciences Research Laboratory in Fargo, North Dakota; FSIS in Athens; and Pennsylvania State University in University Park.
Medina and colleagues developed a method to make a latex chemical agent that hooks, or binds, to bacterial cells in the presence of antibodies against any of the Big Six pathogens. This binding lets food safety scientists verify the presence of any one of the six major non-O157 STECs individually.
To produce antibodies against the STECs, the ERRC researchers injected dead bacteria of individual types into laboratory animals. This activated the animals’ immune systems, which produced the antibodies (IgG) that were collected, purified, and then linked to plastic (polystyrene latex) particles, resulting in a latex-IgG complex.
|
|
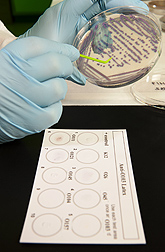
When the researchers mixed a fresh colony of E. coli onto a slide or test card laden with the latex-IgG complex, clumping of the STEC colony was observed instantly. Clumping indicated that the STEC cells were binding to the antibodies, giving a positive result. Flaking, or suspension, indicated bacteria would not bind to the antibodies, giving a negative result.
Through a cooperative partnership with Abraxis LLC, Warminster, Pennsylvania, the technology was tested and validated. In July 2012, commercial test kits based on the ERRC latex-IgG complex and manufactured by Abraxis were rolled out at the International Association for Food Protection conference.
“The IgG is uniformly attached to the surface of microbeads, forming a latex-IgG complex used for test kits,” says Medina. “The IgG on the microbeads acts like a molecular hook, and when the beads bump into the bacteria, they capture the bacteria.”
The ERRC technology is a key component of commercialized test kits, which are now being used by FSIS laboratories that receive meat samples collected from meat processing plants for analysis, according to Cray, a coauthor of the study.
Diagnostic polymerase chain reaction-based tests are first used for preliminary identification of individual STEC groups in the meat samples. Then the test kits are used to confirm the presence of live colonies of STECs belonging to the specific group. The published method for making the latex-IgG complex is available on the FSIS website.
Thanks to the collaboration of FSIS and ARS, and a prompt response from ERRC scientists, a fast, effective detection method for identifying the top six non-O157 STECs on meat samples is available for food safety efforts that help protect consumers.—By Rosalie Marion Bliss, Agricultural Research Service Information Staff.
This research is part of Food Safety, an ARS national program (#108) described at www.nps.ars.usda.gov.
To reach scientists mentioned in this article, contact Rosalie Bliss, USDA-ARS Information Staff, 5601 Sunnyside Ave., Beltsville, MD 20705-5128; (301) 504-4318.
"Detecting Deadly Colonies of E. coli" was published in the July 2013 issue of Agricultural Research magazine.